Biomolecular Simulations SM 2025
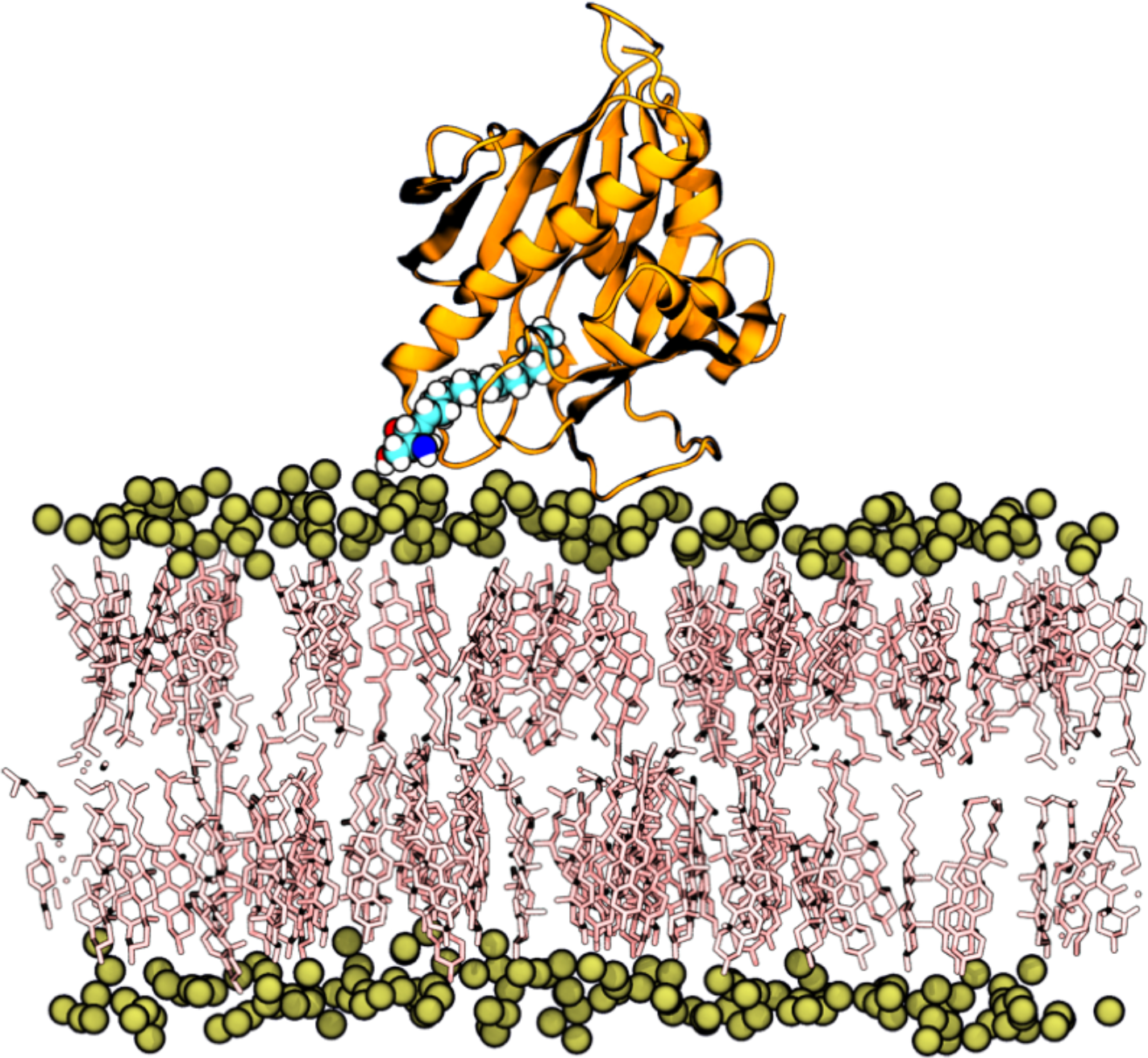
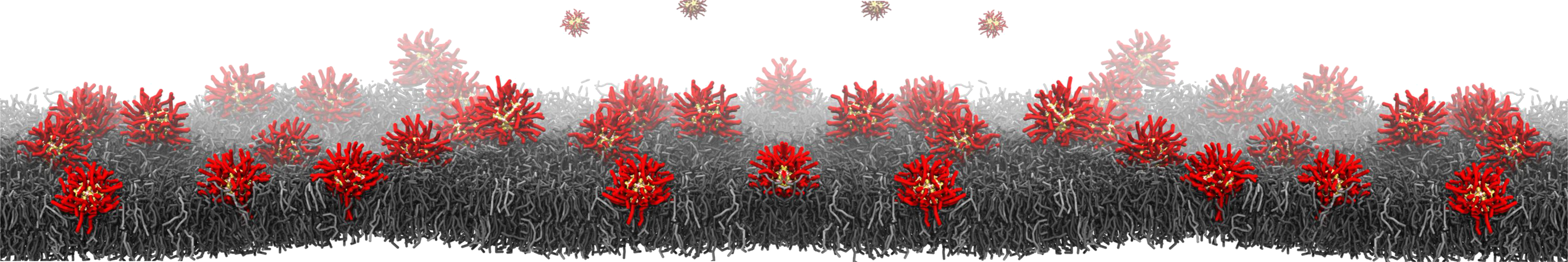
How do proteins interact with each other, how do proteins recognize and bind to specific lipids in complex membranes, and what are the underlying forces at play? These questions can be explored through the lens of the ultimate microscope: computer simulations. This course explores computational methods for studying biomolecular systems' structure, dynamics, and mechanics, including proteins and lipids, at various scales. It aims to equip students with a deep understanding of molecular simulation techniques' principles, capabilities, and limitations. A particular emphasis is placed on Atomistic and coarse-grained Molecular Dynamics simulations, given their extensive use in studying protein-protein interactions, protein-membrane interactions, lipid and protein sorting, and membrane remodeling.This course alternates between theoretical lectures and practical computer tutorials led by Dr. Fabio Lolicato. The hands-on sessions enable advanced Bachelor and interested Ph.D. students to apply the principles of running and analyzing computer simulations of biological matter, enhancing their understanding of molecular simulations.
Copyright © 2023 - Fabio Lolicato